RStudio’s multiple cursors rule!
(TL;DR: Come on. This is pretty short. Productivity level up by harnessing the power of RStudio!)

Motivation
Say we’re at work and we’ve received some data in Excel.
Let’s say that this Excel workbook contains a subset of a broader dataset. For example, maybe our Excel file contains:
- a list of customer ID numbers that we need to look into, or
- a list of dates where our data looks weird and we need to find out what has gone wrong.
Maybe the broader dataset is located within a database. Maybe we have a larger dataset within R itself. In any case, we want to use this subset of data to filter a broader dataset and get more info that might help us in our data analysis adventure!
What I’ve seen a lot of people do in Excel (including myself!)
Let’s proceed with a date and time related example given my recent obsession with time zones. We’ve been given some file of dates to look into and it looks like this:
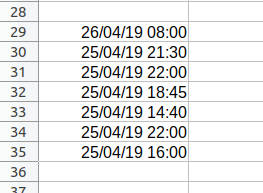
We want to collapse these into quoted, comma-separated strings that we’ll pass into the c() function. This will give us a character vector. Once we have a character vector, we want to convert these into POSIXct values using the power of vectorisation!
In the past, I would have embarassingly done something like this to enclose each date in single quotes with a trailing comma at the end of each line:
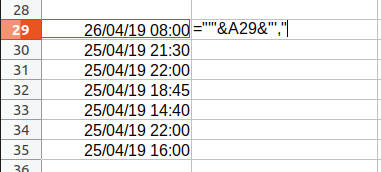
But this requires far too many keystrokes! My new mechanical keyboard with CHERRY MX Brown keyswitches are rated for only 50 million keystrokes per switch. I must be frugal with my keystrokes!
Lazy is good - let’s try this instead
Let’s harness the power of RStudio.
-
We’ll copy the values in the spreadsheet column and paste them into a new script in RStudio:
-
If we’re using a PC keyboard, let’s hold down
Altand click and drag the cursor down the left-hand side of each row until we reach the last line. If we’re using a Mac, we’ll have to do some Googling…but I can only assume that we should use theoptionkey instead of theAltkey!
-
Let’s highlight all of our pasted text row-wise. Let’s hold down
Alt+Shift, then press theright arrow.
-
We’ll quote them lines using another shortcut -
Shift+'(Shiftand thequotation mark key):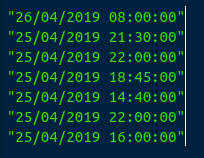
-
While our cursor is still across all lines of the last column, we’ll
press the comma key: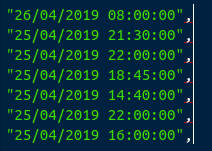
-
Once we’ve removed the annoying trailing comma on the last line, we’ve got ourselves some quoted, comma-separated strings!
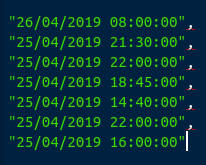
Now we can pass these into the c() function and continue on with our lives. For example, we now have this:
c("26/04/2019 08:00:00",
"25/04/2019 21:30:00",
"25/04/2019 22:00:00",
"25/04/2019 18:45:00",
"25/04/2019 14:40:00",
"25/04/2019 22:00:00",
"25/04/2019 16:00:00")
## [1] "26/04/2019 08:00:00" "25/04/2019 21:30:00" "25/04/2019 22:00:00"
## [4] "25/04/2019 18:45:00" "25/04/2019 14:40:00" "25/04/2019 22:00:00"
## [7] "25/04/2019 16:00:00"
We assign it to a variable:
date_time_str <- c("26/04/2019 08:00:00",
"25/04/2019 21:30:00",
"25/04/2019 22:00:00",
"25/04/2019 18:45:00",
"25/04/2019 14:40:00",
"25/04/2019 22:00:00",
"25/04/2019 16:00:00")
We use the dmy_hms() function from lubridate to convert each element of our vector into POSIXct objects. Since we’re in Sydney, Australia, we’ll assign each the ‘Australia/Sydney’ time zone:
library(lubridate)
date_time_values <- dmy_hms(date_time_str, tz='Australia/Sydney')
print(date_time_values)
## [1] "2019-04-26 08:00:00 AEST" "2019-04-25 21:30:00 AEST"
## [3] "2019-04-25 22:00:00 AEST" "2019-04-25 18:45:00 AEST"
## [5] "2019-04-25 14:40:00 AEST" "2019-04-25 22:00:00 AEST"
## [7] "2019-04-25 16:00:00 AEST"
Hooray!
And since we’re in Sydney, Australia…
We’ll end with this:
“Bloody brilliant!”
Justin